|
Lysine Deacetylases (KDACs)
Due to pandemic drug resistance in the treatment of parasitic infections, there is an urgent need to identify novel drug targets and their associated drug compounds. Although "drug repurposing", i.e. the application of known drugs and compounds to new indications such as infectious diseases, provides a cost effective approach in the development of novel therapeutics, selectivity is one of the major obstacles to overcome in getting such compounds into clinical trials as anti-parasitic drugs. Using the lysine deacetylases (KDACs) as an example, we explored the activities of a panel of known inhibitors against the KDAC targets in a range of parasitic organisms. The computational study of their binding modes to the targets (by docking the compounds to the homology models within different organisms in comparison with the human proteins) helps to rationalize the different activities observed and provide insight on the optimization of lead compounds to improve selectivity. Our work provides support of “drug repurposing” in the treatment of parasitic diseases, and demonstrates the necessity of optimizing these leads for the ultimate goal of preparing them for clinical use.
|
Citation:
Wang Q, Rosa BA, Nare B, et al. Targeting Lysine Deacetylases (KDACs) in Parasites. Geary TG, ed. PLoS Neglected Tropical Diseases. 2015;9(9):e0004026. doi:10.1371/journal.pntd.0004026. |
|
|
Highlights:
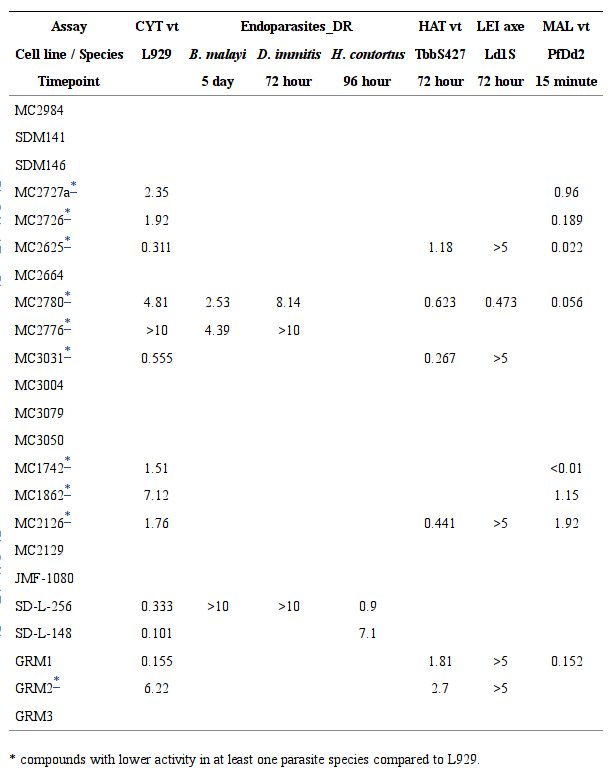
Compound screening in host cells and parasites.
Only measured activities were reported in the table, in the units of nM. The vertical line in the table separates host and parasites. Abbreviation used: CYT vt: cytotoxicity viability assay; Endoparasites_DR: endoparasites dose response assay; HAT vt: Human African trypanosoma viability assay; LEI axe: Leishmania axenic amastigote assay; MAL vt: Malaria viability assay. L929: L929 mouse fibroblast; TbbS427: T. brucei strain S427; Ld1S: L. donovani strain MHOM/SD/00/LS; PfDd2: P. falciparum 3D7 strain.
|
KDAC proteins inferred for the parasitic species within protein families.
Those with the same superscripts (A, B, C) are clustered within the same family. Color codes provide the number of total proteins from each species within an orthologous protein family.

|
Conclusions
A systematic study of all KDAC proteins within parasitic species from protists to nematodes as novel drug targets is reported. Although much work is still required to elucidate their functions and essential nature of these KDACs in parasites, preliminary compound screening suggests that they are inhibited by known host KDAC ligands; such compounds also showed different degrees of selectivity within the parasitic panel. Molecular modeling in combination with genomic profiling offered insight in the mechanism of selectivity and suggested future directions for selectively targeting parasitic lysine deactylases.
|
|
|
Supplemental files:
|
| Image: pntd.0004026.s001.tiff - Compound MC2780 docked to the KDAC1 protein of P. falciparum |
| Image: pntd.0004026.s002.tif - Sequence alignments of all the KDAC1 proteins of parasitic species within the family, along with the sequence of H. sapiens |
| Table: pntd.0004026.s003.xlsx - Sequence identity and similarity between each parasite species target and the template (human) structure, as well as the RMSD values in each step of the modeling process |
| File: pntd.0004026.s004.zip - A compressed file containing protein data bank-format file for the top 5 scoring poses of docked ligands to the KDAC1 structures for the 4 species (H. sapiens, B. malayi, L. donovani and P. falciparum) in pdb format |
|
|
|




