|
|
HelmCoP - a database for drug discovery and protein annotation
|
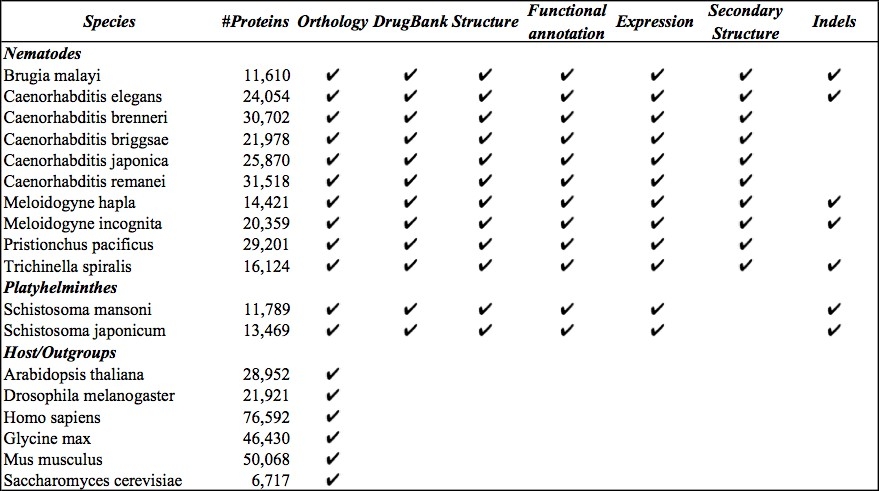
Approximately 85% of the burden from neglected tropical diseases result from helminth infections (nematodes and platyhelminthes). Parasitic helminthes infect over 2 billion, exerting a high collective burden that rivals high-mortality conditions such as AIDS or malaria, and cause devastation to crops and livestock. The challenges to improve control of parasitic helminth infections are multi-fold and no single category of approaches will meet them all. New information such as helminth genomics, functional genomics and proteomics coupled with innovative bioinformatic approaches provide fundamental molecular information about these parasites, accelerating both basic research as well as development of effective diagnostics, vaccines and new drugs. To facilitate such studies we have developed an online resource, HelmCoP (Helminth Control and Prevention), built by integrating functional, structural and comparative genomic data from plant, animal and human helminthes to enable researchers to develop strategies for drug, vaccine and pesticide prioritization, while also providing a useful comparative genomics platform. HelmCoP encompasses genomic data from several hosts, including model organisms, along with a comprehensive suite of structural and functional annotations, to assist in comparative analyses and to study host-parasite interactions. The HelmCoP interface, with a sophisticated query engine as a backbone, allows users to search for multi-factorial combinations of properties and serves readily accessible information that will assist in the identification of various genes of interest.
|
Publication Link HelmCoP: An Online Resource for Helminth Functional Genomics and Drug and Vaccine Targets Prioritization. Sahar Abubucker, John Martin, Christina M. Taylor, and Makedonka Mitreva, PLoS One. 2011; 6(7): e21832.
|
Search by gene:
Search our gene database by using a number of possible keywords. Search by gene name if known, or enter an EC number, KO id, or ontology term to be given a list of all genes in our database associated with that feature.
|
Search by ortholog:
Search our gene database by using a number of possible keywords to define lists of orthologs with at least 1 member meeting those constraints. All gene members of the defined orthologs that meet constraints will be listed.
|
HelmCoP BLAST:
Search your nucleotide or protein sequences against the HelmCoP organisms protein databases. WU-BLASTN or WU-BLASTX alignments will be reported to you via email.
|
|
For more details, please see Frequently Asked Questions. Please contact us at nematode@genome.wustl.edu if you have any questions.
Please use the Firefox browser for the best viewing experience.
|
|
|




